Structure of the Complex of an Iminopyridinedione Protein Tyrosine Phosphatase 4A3 Phosphatase Inhibitor with Human Serum Albumin.
Czub, M.P., Boulton, A.M., Rastelli, E.J., Tasker, N.R., Maskrey, T.S., Blanco, I.K., McQueeney, K.E., Bushweller, J.H., Minor, W., Wipf, P., Sharlow, E.R., Lazo, J.S.(2020) Mol Pharmacol 98: 648-657
- PubMed: 32978326
- DOI: https://doi.org/10.1124/molpharm.120.000131
- Primary Citation of Related Structures:
6WUW - PubMed Abstract:
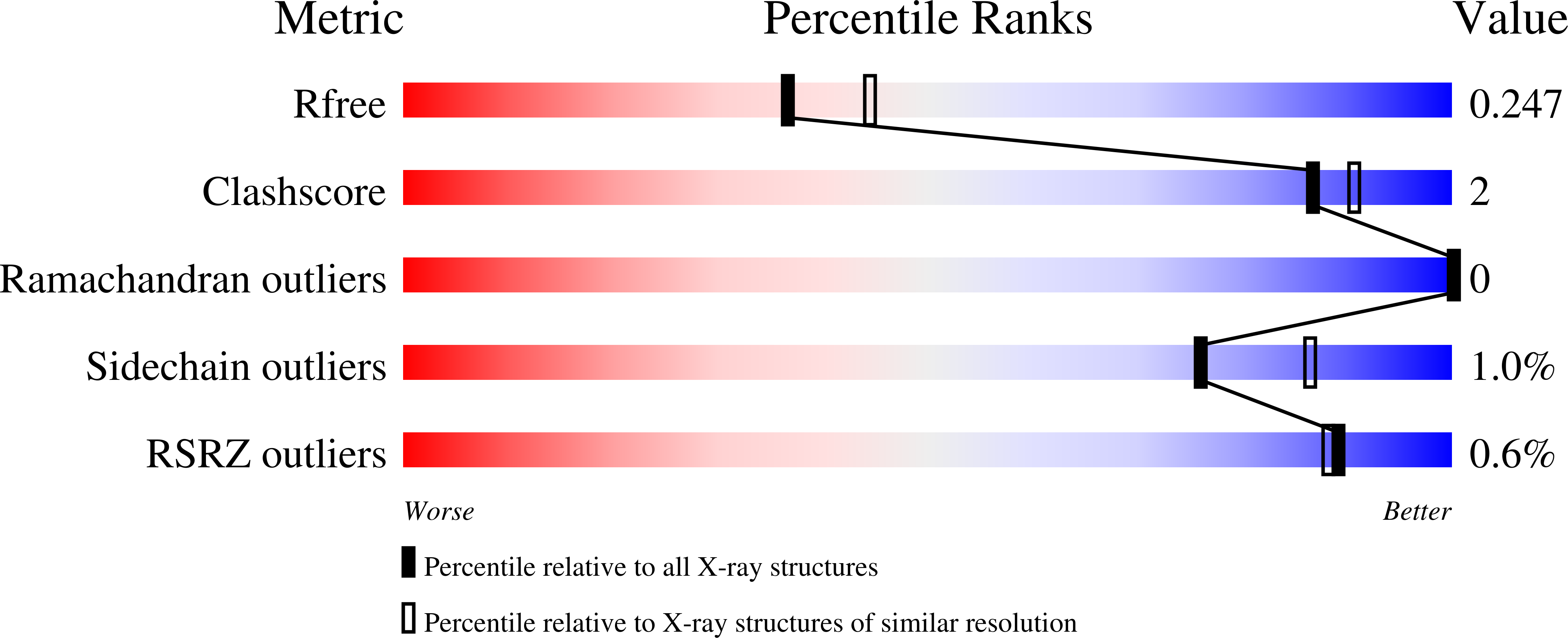
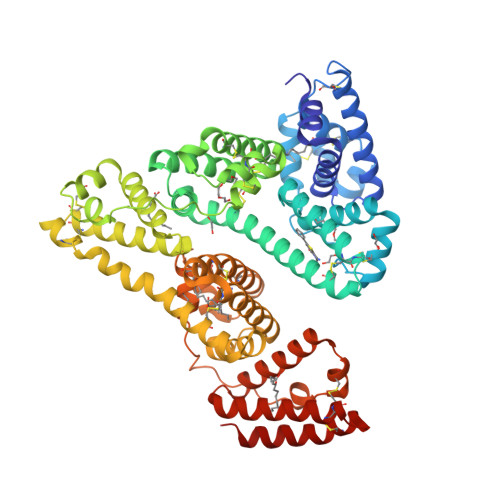
Protein tyrosine phosphatase (PTP) 4A3 is frequently overexpressed in human solid tumors and hematologic malignancies and is associated with tumor cell invasion, metastasis, and a poor patient prognosis. Several potent, selective, and allosteric small molecule inhibitors of PTP4A3 were recently identified. A lead compound in the series, JMS-053 (7-imino-2-phenylthieno[3,2- c ]pyridine-4,6(5 H ,7 H )-dione), has a long plasma half-life (∼ 24 hours) in mice, suggesting possible binding to serum components. We confirmed by isothermal titration calorimetry that JMS-053 binds to human serum albumin. A single JMS-053 binding site was identified by X-ray crystallography in human serum albumin at drug site 3, which is also known as subdomain IB. The binding of JMS-053 to human serum albumin, however, did not markedly alter the overall albumin structure. In the presence of serum albumin, the potency of JMS-053 as an in vitro inhibitor of PTP4A3 and human A2780 ovarian cancer cell growth was reduced. The reversible binding of JMS-053 to serum albumin may serve to increase JMS-053's plasma half-life and thus extend the delivery of the compound to tumors. SIGNIFICANCE STATEMENT: X-ray crystallography revealed that a potent, reversible, first-in-class small molecule inhibitor of the oncogenic phosphatase protein tyrosine phosphatase 4A3 binds to at least one site on human serum albumin, which is likely to extend the compound's plasma half-life and thus assist in drug delivery into tumors.
Organizational Affiliation:
Departments of Molecular Physiology and Biological Physics (M.P.C., A.M.B., J.H.B., W.M.) and Pharmacology (K.E.M., E.R.S., J.S.L.) and Center for Structural Genomics of Infectious Diseases (CSGID) (M.P.C., W.M.), University of Virginia, Charlottesville, Virginia; Department of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania (E.J.R., N.R.T., T.S.M., P.W.); and KeViRx, Inc., Charlottesville, Virginia (I.K.B., E.R.S., J.S.L.).